Welcome to SLAM!
SLAM is the first computational method for structure-aware lysine β-hydroxybutyrylation prediction, designed with a hybrid deep learning neural Networks combining Structure and LAnguage-Model constraints (SLAM).

A geometric deep learning-based method for structure-aware lysine β-hydroxybutyrylation prediction
SLAM is the first computational method for structure-aware lysine β-hydroxybutyrylation prediction, designed with a hybrid deep learning neural Networks combining Structure and LAnguage-Model constraints (SLAM).

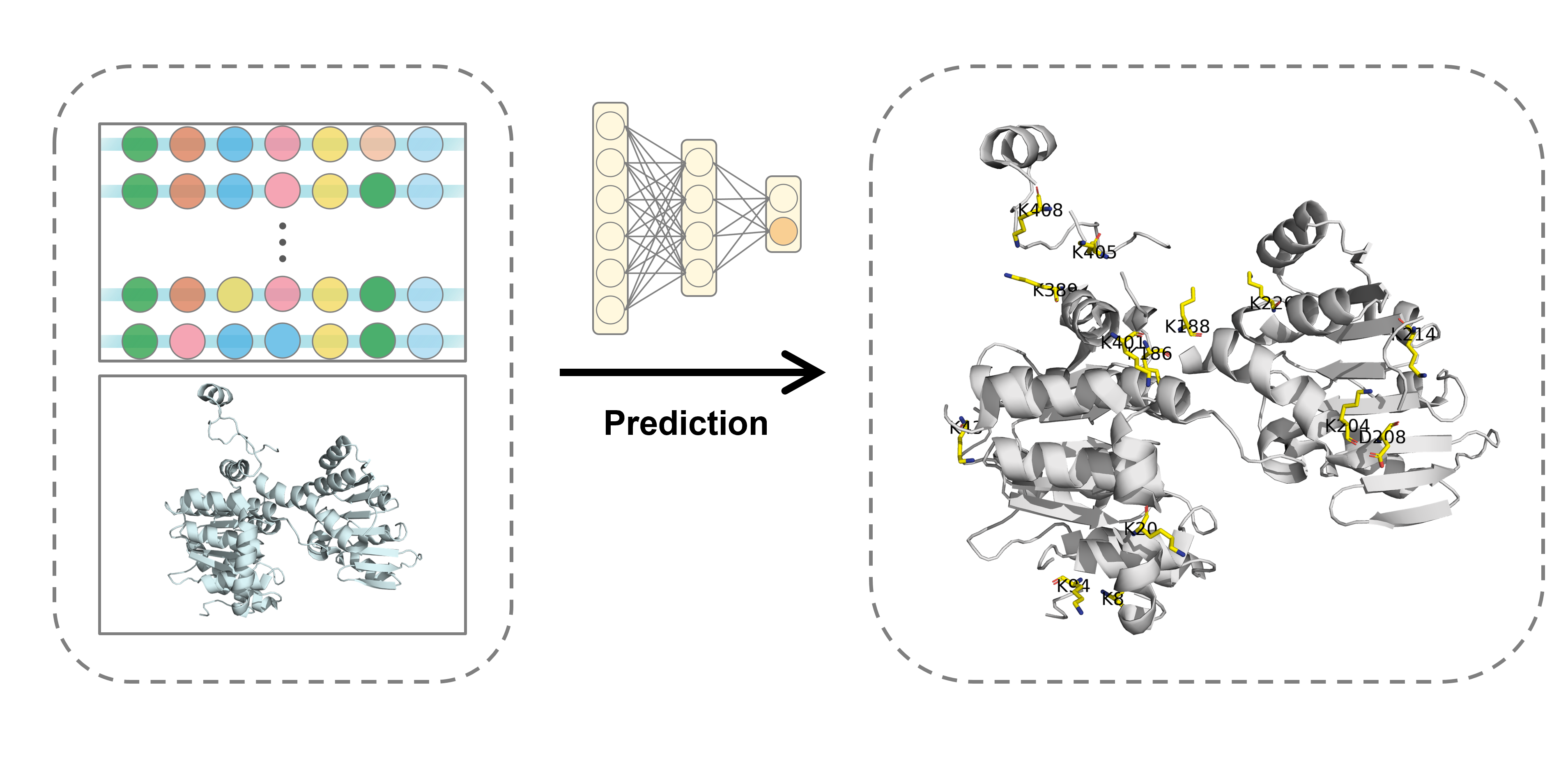
Post-translational modifications (PTM) in proteins expand the functional diversity that are vital for their functions and stability. As a newly-reported PTM, lysine β-hydroxybutyrylation (Kbhb) presents a new avenue to regulate chromatin and diverse functions. Therefore, accurate and efficient prediction of Kbhb sites is imperative. However, the current experimental methods for identifying PTM sites are often expensive and time-consuming. Up to now, there is no computational method for Kbhb sites detection. To this end, we present the first deep learning-based method, termed SLAM, to identify lysine β-hydroxybutyrylation in silico. The performance of SLAM is evaluated on both 5-fold cross-validation and independent test sets for general and three species, achieving 0.876, 0.873, 0.856 and 0.884 in terms of AUC values, respectively on the general and species-specific independent test sets. Furthermore, we found that the species-specific prediction is important for organism with large-scale data, and general prediction still serves as the best approach in species with small-sized dataset. With various in silico experiments, it is confirmed that structure, and information obtained from protein language model and handcrafted features are key contributors to a robust and accurate predictor. As one example, we predicted the potential Kbhb sites in human S-adenosyl-L-homocysteine hydrolase, which is in agreement with experimentally-verified Kbhb sites. Taken together, our method could enable accurate and efficient discovery of novel Kbhb sites that are crucial for the function and stability of proteins and could be applied in the structure-guided identification of other important PTMs. The source code of SLAM is freely accessible at SLAM.

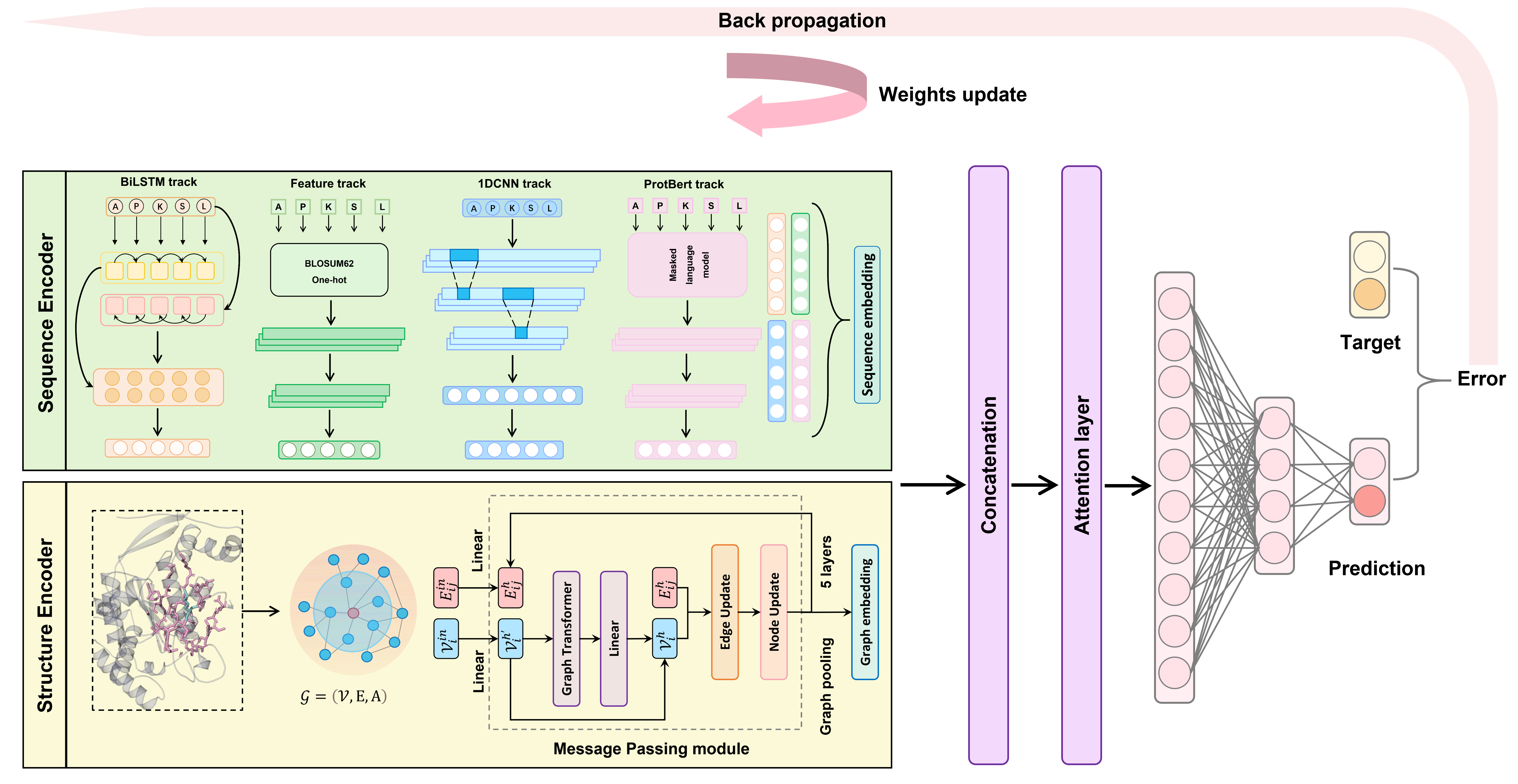
We present a hybrid deep learning neural Networks combining Structure and LAnguage-Model constraints (SLAM), for species-specific and general protein lysine β-hydroxybutyrylation site prediction. The developed geometric deep learning framework includes 1) a multi-track encoder module to concurrently embed the protein structure and sequence features into a latent representation; 2) a decoder layer consisting of an attention layer and a multi-layer perceptron followed with a sigmoid function for downstream classification. The sequence encoder is designed as hybrid deep learning neural networks to learn dependencies between residues with two-track feature encoders and two-track adaptive encoders. Adaptive encoders enable learn-from-data for SLAM by using learnable word embeddings, and feature encoders provide expert-level information and evolutionary constraints extracted from protein language model. For structure encoder, a multi-layer graph neural network (GNN) is implemented to capture high-level residue relationships considering geometry.